If you’ve ever looked at a Makefile in a python or R repository chances are that it contained a collection of useful shell commands (make test -> runs all the unit tests, make lint -> runs automatic formatting and linting, etc.). That’s a perfectly good use of make, and if that’s what you’re after then
here’s a good guide for how to set that up. However, the original reason why make was made was to run shell commands, which might depend on other commands being run first, in the right order. In
1976 when Stuart Feldman created make those shell commands were compiling C-programs, but nothing is stopping you from using make to set up simple data pipelines instead. And there are a couple of good reasons why you would want to use make for this purpose:
makeis everywhere. Well, maybe not on Windows (but it’s easy to install), but on Linux and MacOSmakecomes installed out of the box.makeallows you to define pipelines that have multiple steps and complex dependencies (this needs to run before that, but after this, etc.), and figures out what step needs to be rerun and executes them in the correct order.makeis language agnostic and allows you to mix pipelines with Python code, Jupyter notebooks, R code, shell scripts, etc.
Here I’ll give you a handy template and some tips for building a data pipeline using python and make. But first, let’s look at an example data pipeline without make.
A python data pipeline, but without make
As an example, let’s take a minimal, somewhat contrived, data pipeline resulting in an analysis of Pokemon data. This data pipeline could easily fit into a single notebook but is here a stand-in for a more complex data pipeline. In this data pipeline, we’ll
- Download and clean up a file with Pokemon statistics.
- Download files with English and German Pokemon names, and join them together. (Why? I guess we want to cater to the substantial German audience of Pokemon analytics…)
- Render a Jupyter notebook analysis of these data as an HTML document.
You can find the full data pipeline code here without output, here with all output, and here’s the final Pokemon “analysis”. But the important part is not the code. What’s important is the problem we have now: As we’ve put each step of this analysis into its own script, we now have several scripts we need to run, ideally in the right order. One way to do that is to stick it all into a shell script, say, run-analysis.sh:
# Downloading and cleaning all the data
python pokemon-stats/01_download.py
python pokemon-stats/02_clean.py
python pokemon-names/01_download-en.py
python pokemon-names/02_download-de.py
python pokemon-names/03_combine.py
# Rendering the final report to html
jupyter nbconvert --execute quick-pokemon-analysis.ipynb --to html
For our minimal example, this will work quite well. But if our data pipeline would grow we would start to get three problems:
- Even if we just change something at the end of the pipeline (say
quick-pokemon-analysis.ipynb) we’ll have to run the whole pipeline every time. - We have to manually make sure that the python scripts are listed in the right order. Maybe easy when there are six scripts, but it can quickly become unmanageable.
- It’s not clear in the shell script what the inputs and output are of each python script.
A python data pipeline, now with make
Switching out the shell script for a Makefile solves these three problems as the Makefile will include the whole dependency graph of the pipeline. That is, what files does each output in the pipeline depend on? For our simple example, the dependency graph looks like this:
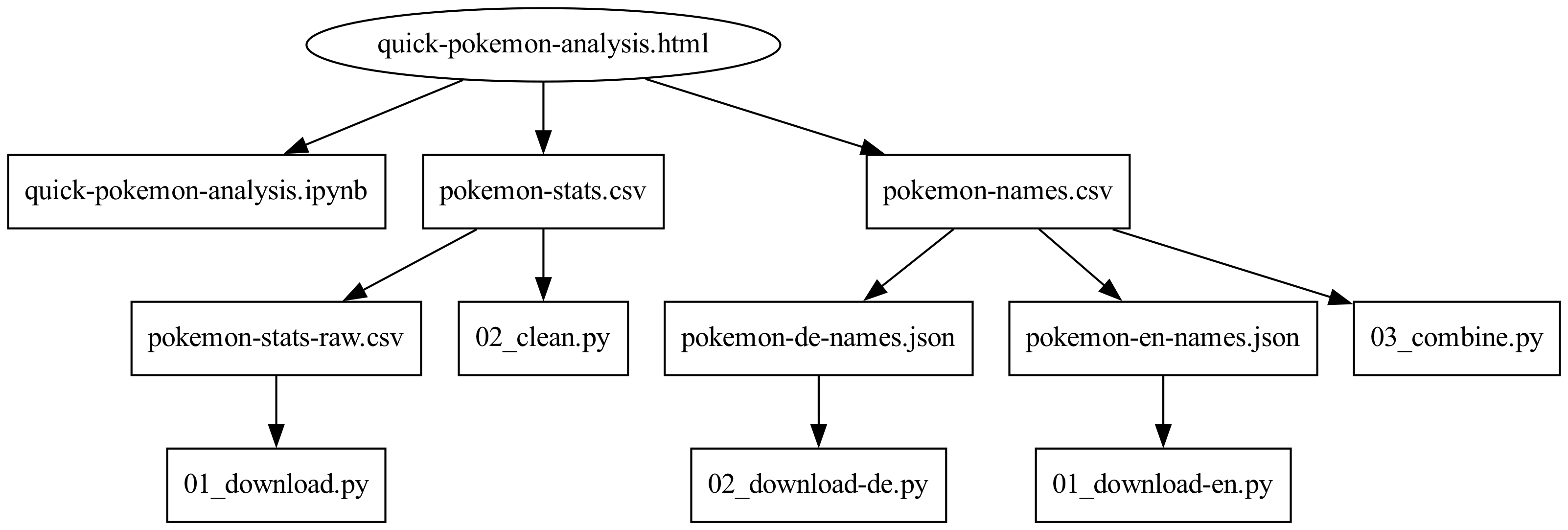
Arrows here point to the dependencies so, for example, if either 02_clean.py or pokemon-stats-raw.csv changes we would need to regenerate pokemon-stats.csv. Using this dependency graph make will figure out in what order things need to run and what needs to be re-run (there’s no need to re-run pokemon-stats/01_download.py if we’ve only made changes to quick-pokemon-analysis.ipynb, say).
The Makefile syntax for defining dependency graphs is a little bit unusual, but a Makefile rule looks like this:
the_output_file: the_first_dependency the_second_dependency the_third_dependency etc.
shell_command_generating_the_output_file
For pokemon-stats.csv in our pipeline the Makefile rule would look like this:
pokemon-stats/pokemon-stats.csv: pokemon-stats/02_clean.py pokemon-stats/pokemon-stats-raw.csv
python pokemon-stats/02_clean.py
Big Gotcha: The indentation before the shell command needs to be there, and needs to be a tab. Not two spaces, not four spaces, an actual tab character. Accidentally putting spaces there instead of a tab is a very common problem.
We’re now ready to write up the whole dependency graph as a Makefile, but there are a couple of tricks we can use to make this rule prettier and less verbose:
- We can assign the root folder
pokemon-statsto the variablep(as in path) so that we don’t need to repeat it everywhere. This variable can then be inserted by putting a$in from, like this:$p. - We can separate the two dependencies on different lines using
\which “continues the line” on the next row. - We can use the magic
$<variable, which always refers to the first dependency (here02_clean.py), to avoid having to repeat the name of the script.
With this, the rule would become:
p := pokemon-stats
$p/pokemon-stats.csv: $p/02_clean.py \
$p/pokemon-stats-raw.csv
python $<
(Note that the second dependency, $p/pokemon-stats-raw.csv, has leading spaces while python $< still needs a leading tab)
The full dependency graph as a Makefile (traditionally also given the file name Makefile) would then be:
quick-pokemon-analysis.html: quick-pokemon-analysis.ipynb \
pokemon-stats/pokemon-stats.csv \
pokemon-names/pokemon-names.csv
jupyter nbconvert --execute $< --to html
p := pokemon-stats
$p/pokemon-stats-raw.csv: $p/01_download.py
python $<
$p/pokemon-stats.csv: $p/02_clean.py \
$p/pokemon-stats-raw.csv
python $<
p := pokemon-names
$p/pokemon-en-names.json: $p/01_download-en.py
python $<
$p/pokemon-de-names.json: $p/02_download-de.py
python $<
$p/pokemon-names.csv: $p/03_combine.py \
$p/pokemon-en-names.json \
$p/pokemon-de-names.json
python $<
So, slightly longer than the run-analysis.sh shell script we started out with, but now all the dependencies are there. Note that the order of the rules doesn’t matter except that make, by default, targets making the first output it encounters. Here it’s quick-pokemon-analysis.html, which makes sense, as that’s the ultimate output of the pipeline. To run this you open up a terminal, navigate to the folder of the Makefile and run make, which will give you the following output:
> make
python pokemon-stats/01_download.py
python pokemon-stats/02_clean.py
python pokemon-names/01_download-en.py
python pokemon-names/02_download-de.py
python pokemon-names/03_combine.py
jupyter nbconvert --execute quick-pokemon-analysis.ipynb --to html
[NbConvertApp] Converting notebook quick-pokemon-analysis.ipynb to html
[NbConvertApp] Writing 609122 bytes to quick-pokemon-analysis.html
From this, we can see that make figured out the correct order to run things. Now the exciting thing is: what happens if we run make again?
> make
make: `quick-pokemon-analysis.html' is up to date.
Nothing happens! make has figured out that, because we haven’t changed any of the dependencies since last time, nothing needs to be rerun. What if we make an update to, say, pokemon-names/02_download-de.py and run make again?
> make
python pokemon-names/02_download-de.py
python pokemon-names/03_combine.py
jupyter nbconvert --execute quick-pokemon-analysis.ipynb --to html
[NbConvertApp] Converting notebook quick-pokemon-analysis.ipynb to html
[NbConvertApp] Writing 609122 bytes to quick-pokemon-analysis.html
Then make only reruns what’s needed, according to the dependency graph, and nothing more. That’s the beauty of make!
A python Makefile template
make is an old and deep piece of software, there are many things to learn and also many ways things can go wrong. But here’s a Makefile template that gets you started with using make for python data pipelines.
# Some reasonable makefile settings
# Always use the bash shell instead of the default shell which might be zsh, fish, etc.
SHELL := bash
# Flags to put bash into "strict" mode
.SHELLFLAGS := -eu -o pipefail -c
MAKEFLAGS += --warn-undefined-variables
# We're not compiling C++, so no need for the many built-in rules
MAKEFLAGS += --no-builtin-rules
p := some/path
$p/an-output.csv: $p/a-script.py \
$p/a-dependency.csv \
$p/another-dependency.csv
python $<
$p/another-output.csv: $p/another-script.py \
$p/an-output.csv
python $<
# ---------------------------------
# An explanation of a recurring pattern in this Makefile. To avoid repeating the same
# path multiple times, for scripts that live in folders, for example:
# ---------------------------------
# some/path/an-output.csv: some/path/a-script.py \
# some/path/a-dependency.csv \
# some/path/another-dependency.csv
# python some/path/a-script.py
# ---------------------------------
# We write the above like this instead:
# ---------------------------------
# p := some/path
# $p/an-output.csv: $p/a-script.py \
# $p/a-dependency.csv \
# $p/another-dependency.csv
# python $<
# ---------------------------------
# Some gotchas here:
# * The variable p (as in Path) can be used as $p, but this only works for
# one-character variables, for longer variables parentheses are $(needed).
# * As p might be redefined througout the script the recipe *can't* be
# python $p/a-script.py. As recipes are resolved after the Makefile has been parsed
# all $p in the recipes will be set to the last value assigned to p.
# To avoid this we use the "automatic" variable $< which will be replaced
# by the first pre-requisite. Therefore, the first pre-requisite of every
# rule needs to be the python script, otherwise $< will point to a CSV-file or similar.
Some last tips/warnings when using make for setting up data pipelines:
-
The official
makedocumentation is great, albeit fairly focused on compiling C programs. - Don’t rely on the latest
makefeatures. Chances are that themakethat comes with your OS is fairly old, especially if you are on a Mac (which might be perpetually stuck on version 3.82). - There’s no single good way to state that a python script generates several output files, but there are many half-good ways.
- If you want to split up your
Makefileinto several makefile “modules”, think again. This is very tricky to do, and if you have such a complex pipeline, thenmakeis likely not the right tool for the job to begin with. - Make is great for small data pipelines because it’s an old, stable tool that’s already installed everywhere. However, if you are dealing with a complex data pipeline there are much better tools out there such as
Snakemake or
targetsif you’re in an R setting. - Finally, if you’re getting weird errors, like
*** missing separator. Stop., then you probably have spaces in front of a command where you should have a tab.